Multi‑Modal Data Integration
Predict Alzheimer’s disease onset and progression by combining neuroimaging, omics, and clinical features with deep learning algorithms.
Open project →Dr. Minghan Chen · Wake Forest University · Computer Science

Dr. Minghan Chen is an Associate Professor of Computer Science at Wake Forest University. Her research focuses on science-guided machine learning and AI for health and biomedical applications. She welcomes interdisciplinary collaborations and is recruiting motivated students in computational biology and machine learning. Interested applicants should email a brief background statement, CV, and transcripts.
Our group develops robust machine learning and statistical methods to connect multi‑modal biomedical data (omics, imaging) with clinical outcomes. Current directions include: disease trajectory modeling, multi‑omics integration, foundation models for biomedical systems, and trustworthy AI for healthcare.
Predict Alzheimer’s disease onset and progression by combining neuroimaging, omics, and clinical features with deep learning algorithms.
Open project →Design graph neural operators to model tau transport across brain and forecast tau spatio-temporal dynamics under different seeding conditions and biophysical parameters.
Explore the Tool →Develop MAMBAxBrain framework to integrate Mamba with functional connectivity analysis for multi-task fMRI modeling, including brain fingerprinting, cognitive decoding, reaction-time prediction, and schizophrenia diagnosis.
We build AI-based surrogate models to accelerate complex systems biology simulations. These surrogates reduce computational cost while maintaining predictive accuracy for pathway and regulatory network analysis.
Highlights from our students’ research, presentations, and summer workshops.

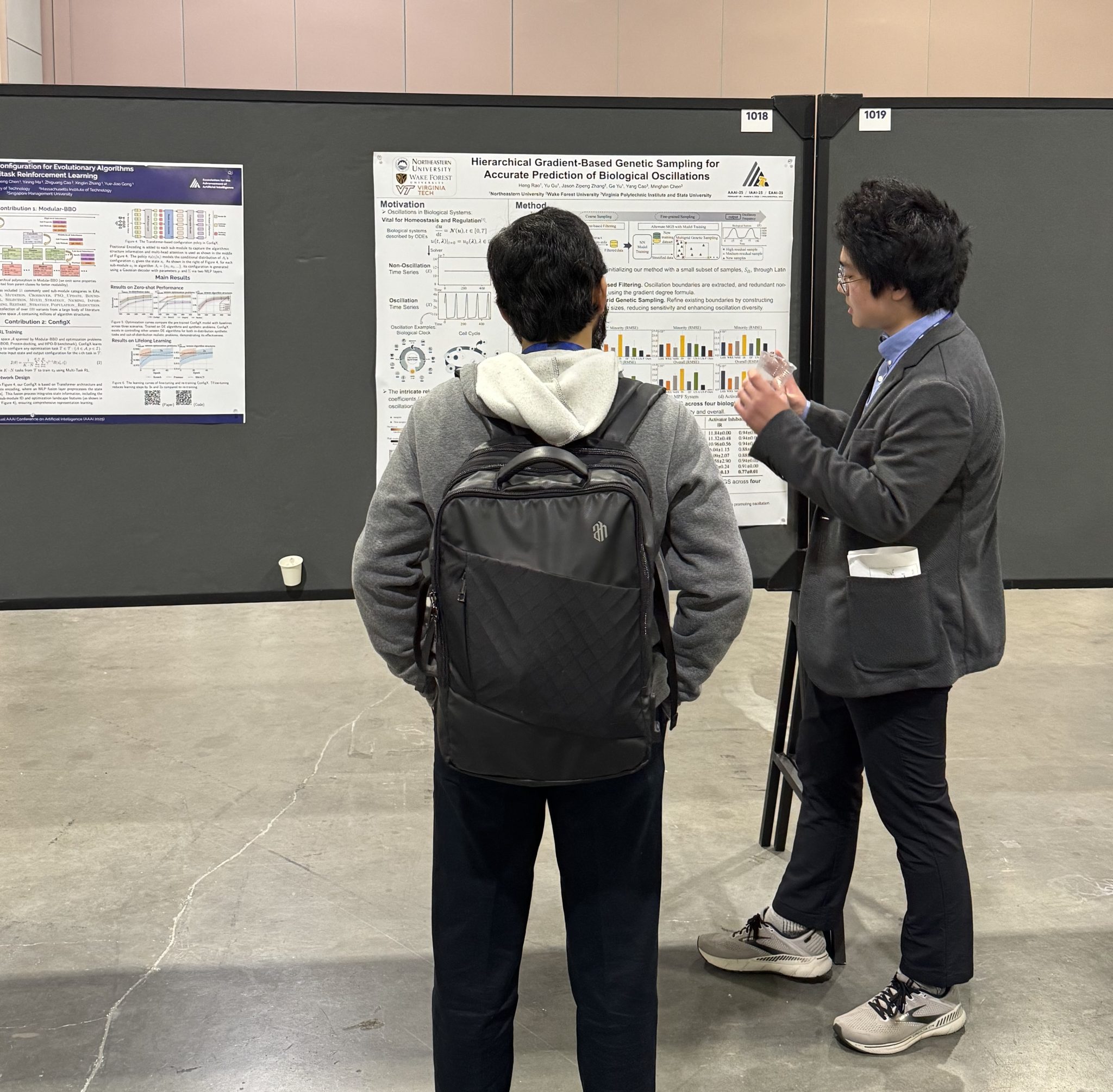

Yang Xiao, 2025 (Computer Science, University of Pennsylvania)
Johnson Wang, 2024 (Computational Biology, Harvard University)
Ruiwen Yang, 2024 (Data Science, University of Pennsylvania)
Selina Zhang, 2023 (Computer Science, Harvard University)
Ziang Xu, 2023 (Computer Science, Columbia University)
Zhanyang Sun, 2023 (Data Science, University of Southern California)
Enze Xu, 2023 (Computer Science, William & Mary College)
Michelle Dai, 2022 (Computational Biology, Harvard University)
Patrick Fan, 2022 (Computational Finance, Carnegie Mellon University)
Laurent Zhang, 2022 (Data Science, New York University)
Bess Morrell, 2022 (Independent Study)
Hao Lin, 2022 (Summer research, Duke University)
David Ding, 2022 (Computer Science, Rice University)
Henry Hollis, 2021 (Biomedical Engineering, Drexel University)
Alex Wei, 2020 (Computational Biology, Harvard University)